
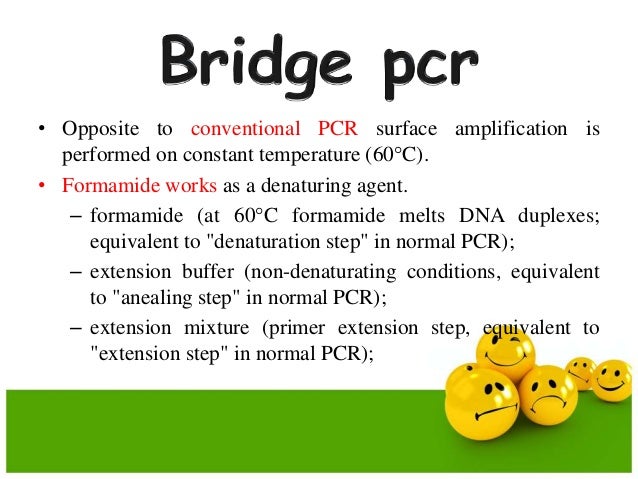
The PCR then denatures the library fragment leading two separate strands, one of which (the reverse strand) anneals to the bead. In order for the sequencing process to be successful, each micro well should contain one bead with one strand of DNA (approximately 15% of micro wells are of this composition). Emulsion PCRĮmulsion oil, beads, PCR mix and the library DNA are mixed to form an emulsion which leads to the formation of micro wells ( Figure 2). Instead, there are several types of amplification process which use PCR to create large numbers of DNA clusters. With enzymatic amplification, phenomena such as 'biasing' and 'duplication' can occur leading to preferential amplification of certain library fragments. Library amplification is required so that the received signal from the sequencer is strong enough to be detected accurately. coli cloning used to isolate and amplify DNA for Sanger sequencing, however, this is at the expense of read length of the fragments. This method of library construction is much faster than the previous labour intensive procedure of colony picking and E. Since these polonies are attached in a planar fashion, the features of the array can be manipulated enzymatically in parallel. In order for sequencing to be successful, the library fragments need to be spatially clustered in PCR colonies or 'polonies' as they are conventionally known, which consist of many copies of a particular library fragment. Library preparation of Next-generation sequencing By removing the phosphate from the sticky end of the adaptor and therefore creating a 5'-OH end instead, the DNA ligase is unable to form a bridge between the two termini ( Figure 1). To prevent this, the chemical structure of DNA is utilised, since ligation takes place between the 3'-OH and 5'-P ends. This could lead to the potential problem of base pairing between molecules and therefore dimer formation. The adaptors enable the sequence to become bound to a complementary counterpart.Īdaptors are synthesised so that one end is 'sticky' whilst the other is 'blunt' (non-cohesive) with the view to joining the blunt end to the blunt ended DNA. Adaptors (short, double-stranded pieces of synthetic DNA) are then ligated to these fragments with the help of DNA ligase, an enzyme that joins DNA strands. Sequencing: DNA is sequenced using one of several different approachesįirstly, DNA is fragmented either enzymatically or by sonication (excitation using ultrasound) to create smaller strands.Amplification: the library is amplified using clonal amplification methods and PCR.Library preparation: libraries are created using random fragmentation of DNA, followed by ligation with custom linkers.Next generation methods of DNA sequencing have three general steps: The genome sequencing projects that took many years with Sanger methods can now be completed in hours with NGS, although with shorter read lengths (the number of bases that are sequenced at a time) and less accuracy. The NGS method uses array-based sequencing which combines the techniques developed in Sanger sequencing to process millions of reactions in parallel, resulting in very high speed and throughput at a reduced cost. The Sanger method required separate steps for sequencing, separation (by electrophoresis) and detection, which made it difficult to automate the sample preparation and it was limited in throughput, scalability and resolution. The genomic strand is fragmented, and the bases in each fragment are identified by emitted signals when the fragments are ligated against a template strand. The principle behind Next Generation Sequencing (NGS) is similar to that of Sanger sequencing, which relies on capillary electrophoresis. Sanger sequencing and Next-generation sequencing Thanks to new sequencing technologies known collectively as Next Generation Sequencing, it is now possible to sequence an entire human genome in a matter of hours. Today, the demand for sequencing is growing exponentially, with large amounts of genomic DNA needing to be analyzed quickly, cheaply, and accurately. The Human Genome Project used Sanger sequencing (albeit heavily optimized), the principal method of DNA sequencing since its invention in the 1970s. The sequencing of the human genome was completed in 2003, after 13 years of international collaboration and investment of USD 3 billion.


 0 kommentar(er)
0 kommentar(er)
